Using Chimera to Create CryoEM Data¶
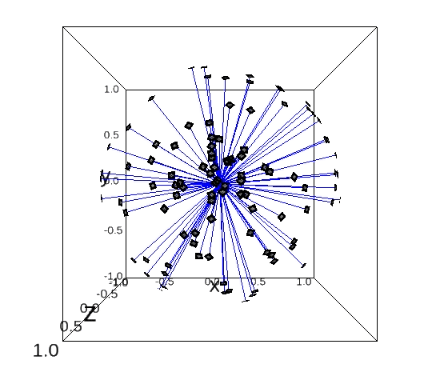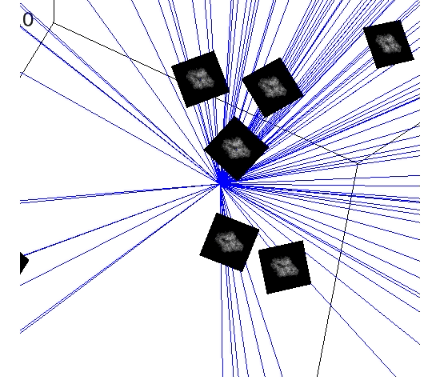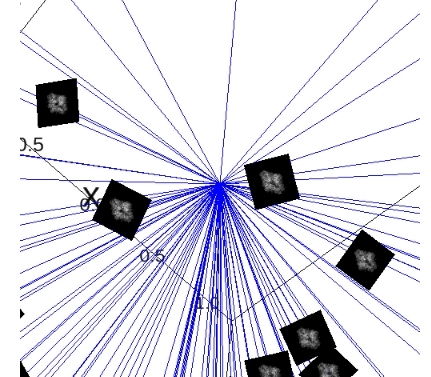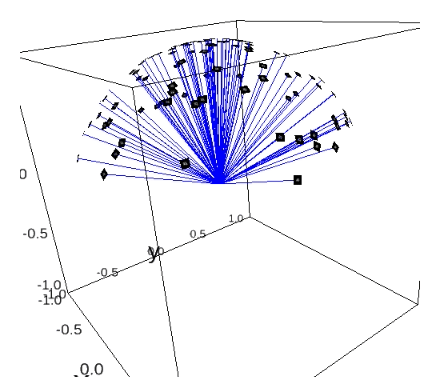
First, we created faithful Cryo-EM data for the neural networks training (including the imaging parameters that you can play with). This is done by generating these ground truths from scratch.
The steps for generating simulation data with chimera:
Chimera software: Download chimera from this page.
Protein in PDB: Find a protein that interests you in the Protein Data Bank (PDB) (let’s say we take beta-galactosidase, with PDB ID 5J0N).
PDB model: Open chimera and import that protein: File -> Fetch By ID… -> Select PDB and type the PDB ID of the protein -> Fetch. Now you will see the 3D structure of the protein.
Create reference map from PDB model: Modify the density of the protein: Tools -> General Controls -> Command Line -> Type:
molmap #0 5(molmap = is command that generates a density map from the specified atom; # is atom specification, i.e. number assigned to the model by default; 5 = resolution which for us does not have sense to work with resolutions above 15, we will have proteins around 2 or 3 Angstrom).Store reference map as MRC: Save to a file: Tools -> Volume Data -> Volume Viewer -> File -> Save map as… -> Give it the
protein_PDB_ID.mrc.
References for more information:
We will be working mainly on this protein: 5j0n (asymmetric).
import nglview as nv
Lambda excision HJ intermediate - 5j0n (asymmetric)¶
More information on the protein can be found here. Following are the atomic model and density map of the protein respectively.
PROTEIN = "5j0n"
view_atomic = nv.show_pdbid(PROTEIN)
view_atomic
view_density_map = nv.show_file(f"../data/{PROTEIN}.mrc")
view_density_map