Generate 2D Projections with Angles¶
From the 3D protein structure, we want to generate 2D projections that were taken at different angles. This is done using the cryoem software tools implemented in this project that rely on Astra Toolbox.
In this notebook we visualize the output of this generator. We have two main spaces we visualize the data:
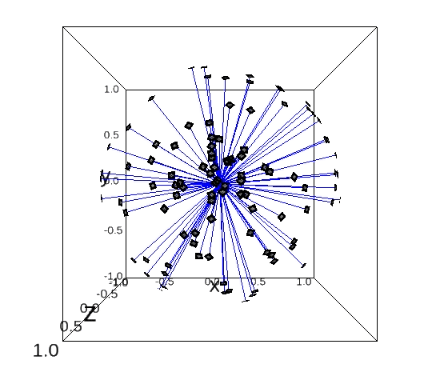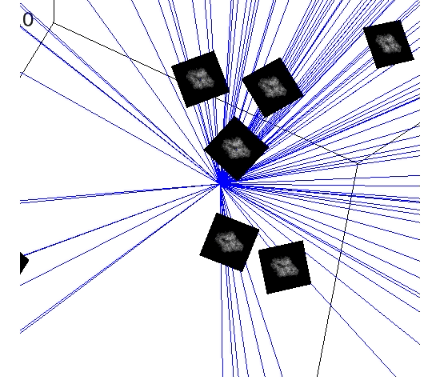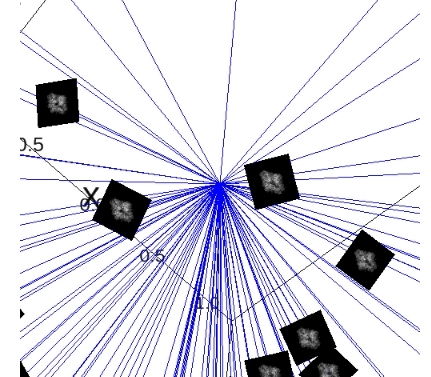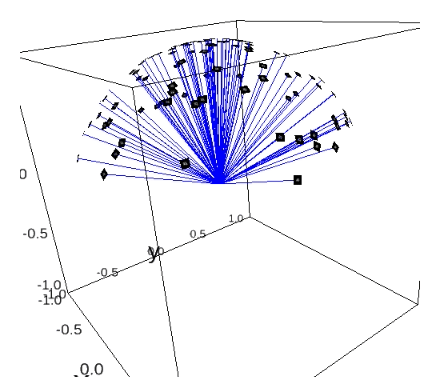
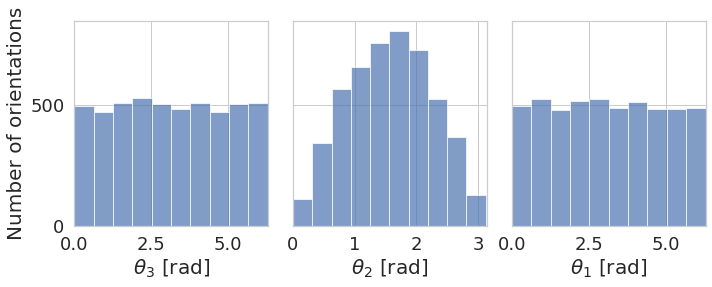
\(\mathbb{S}^2\) space which we use to represent the sampled directions. These directions represents at which angles the ray was sent through the protein. Imagine the protein in the center of the coordinate system and the potential ray positions cover the sphere around that protein with the fixed radius (i.e. only the border of the sphere is filled, the sphere itself is empty). It can also be described as camera positions around the protein. Only two angles \((\theta_2, \theta_1)\) represent the direction and they are used for visualization.
\(\mathbf{SO}(3)\) space with the topology explained here. The possible positions fill the ball with radius that is in the range from \(-\pi\) to \(\pi\). For every point in this ball there is a rotation, with axis through the point and the origin, and rotation angle equal to the distance of the point from the origin. All three angles \((\theta_3, \theta_2, \theta_1)\) are used for the visualization.
Note: The goal of this project is to recover these positions of angles on the sphere only using the projections (images) not knowing the angle at which they were taken.
Following command line will be used to run the script for generating 2D projections with angles. Note: cd ../.. is just locating into the root of this project. So if you run it in the root, you don’t need it.
!python ../../generator.py --help
Warning: To use the exr data format, please install the OpenEXR package following the instructions detailed in the README at github.com/tensorflow/graphics.
Warning: To use the threejs_vizualization, please install the colabtools package following the instructions detailed in the README at github.com/tensorflow/graphics.
usage: generator.py [-h] [--config-file CONFIG_FILE] [--input-file INPUT_FILE]
[--projections-num PROJECTIONS_NUM]
[--angle-shift ANGLE_SHIFT]
[--angle-coverage ANGLE_COVERAGE]
[--angles-gen-mode ANGLES_GEN_MODE]
[--output-file OUTPUT_FILE]
Generator of 2D projections of 3D Cryo-Em volumes Args that start with '--'
(eg. --input-file) can also be set in a config file (protein.config or
specified via --config-file). Config file syntax allows: key=value, flag=true,
stuff=[a,b,c] (for details, see syntax at https://goo.gl/R74nmi). If an arg is
specified in more than one place, then commandline values override config file
values which override defaults.
optional arguments:
-h, --help show this help message and exit
--config-file CONFIG_FILE, -conf CONFIG_FILE
Config file path
--input-file INPUT_FILE, -in INPUT_FILE
Input file of 3D volume (*.mrc format)
--projections-num PROJECTIONS_NUM, -num PROJECTIONS_NUM
Number of 2D projections. Default 5000
--angle-shift ANGLE_SHIFT, -shift ANGLE_SHIFT
Get the start Euler angles that will rotate around
axes Z, Y, Z repsectively. Example usage: -shift 0.0
-shift 0.0 -shift 0.0
--angle-coverage ANGLE_COVERAGE, -cov ANGLE_COVERAGE
The range (size of the interval) of the Euler angles
aroung Z, Y, Z axes respectively. Example usage: -cov
0.0 -cov 0.0 -cov 0.0
--angles-gen-mode ANGLES_GEN_MODE, -ang-gen ANGLES_GEN_MODE
Specify the mode of generating angles [uniform_S2,
uniform_angles]
--output-file OUTPUT_FILE, -out OUTPUT_FILE
Name of output file containing projections with angles
(with the extension)
ROOT_DIR = "../.."
from numba import cuda
cuda.select_device(0)
cuda.close()
import os
import h5py
import numpy as np
import sys
sys.path.append(ROOT_DIR)
from cryoem.projections import RotationMatrix
from cryoem.conversions import euler2quaternion
from cryoem.plots import plot_detector_pixels_with_protein, plot_images, plot_rays, plot_angles_histogram, plot_quaternions_histogram, plot_rotvec, plot_polar_plot, plot_projection, plot_projections
Warning: To use the exr data format, please install the OpenEXR package following the instructions detailed in the README at github.com/tensorflow/graphics.
Warning: To use the threejs_vizualization, please install the colabtools package following the instructions detailed in the README at github.com/tensorflow/graphics.
import tensorflow as tf
from tensorflow.python.client import device_lib
print(device_lib.list_local_devices())
[name: "/device:CPU:0"
device_type: "CPU"
memory_limit: 268435456
locality {
}
incarnation: 6225653293364602262
, name: "/device:XLA_CPU:0"
device_type: "XLA_CPU"
memory_limit: 17179869184
locality {
}
incarnation: 15307045232860424414
physical_device_desc: "device: XLA_CPU device"
, name: "/device:XLA_GPU:0"
device_type: "XLA_GPU"
memory_limit: 17179869184
locality {
}
incarnation: 12847188267404837275
physical_device_desc: "device: XLA_GPU device"
, name: "/device:GPU:0"
device_type: "GPU"
memory_limit: 3188981760
locality {
bus_id: 1
links {
}
}
incarnation: 10639599417925275779
physical_device_desc: "device: 0, name: GeForce GTX 1050 Ti with Max-Q Design, pci bus id: 0000:01:00.0, compute capability: 6.1"
]
Full-Sphere Coverage - Uniform on \(\mathbb{S}^2\) - 5K Projections¶
Generating the 5000 projection images that were taken from angles covering the full sphere around the protein. The sampling on the sphere is done uniformly on \(\mathbb{S}^2\).
To generate simulation data of the protein, you can run the following command.
python generator.py --input-file data/5j0n.mrc -ang-gen uniform_angles -num 5000 -shift 0.0 -shift 0.0 -shift 0.0 -cov 2.0 -cov 1.0 -cov 2.0
Note that the alternative way is just to change the config file protein.config with the modifications that you need. In that case it is sufficient to only run the following command:
python generator.py -conf protein.config
For every projection, we will have 3 angles, each representing the rotation around the corresponding axis. In our case we are dealing with \(Z-Y-Z\) rotations:
Z in \([0, 2\pi]\)
Y in \([0, \pi]\)
Z in \([0, 2\pi]\)
!python ../../generator.py --input-file ../../data/5j0n.mrc --output-file ../../data/5j0n_full_uniformS2.h5 -ang-gen uniform_S2 -num 5000 -shift 0.0 -shift 0.0 -shift 0.0 -cov 2.0 -cov 1.0 -cov 2.0
Warning: To use the exr data format, please install the OpenEXR package following the instructions detailed in the README at github.com/tensorflow/graphics.
Warning: To use the threejs_vizualization, please install the colabtools package following the instructions detailed in the README at github.com/tensorflow/graphics.
----------
Command Line Args: --input-file ../../data/5j0n.mrc --output-file ../../data/5j0n_full_uniformS2.h5 -ang-gen uniform_S2 -num 5000 -shift 0.0 -shift 0.0 -shift 0.0 -cov 2.0 -cov 1.0 -cov 2.0
----------
* Loading the dataset *
Protein: 5j0n
Input filename: ../../data/5j0n.mrc
Output filename: ../../data/5j0n_full_uniformS2.h5
Projections (#): (5000, 116, 116)
Angles (#): (5000, 3)
**********
# half coverage (AngCoverage=0.5)
projections_filename = "../../data/5j0n_full_uniformS2.h5"
# load structures
data_full_5k_uniform = h5py.File(projections_filename, 'r')
angles2 = np.array(data_full_5k_uniform['Angles'], dtype=np.float32)
projections2 = np.array(data_full_5k_uniform['Projections'], dtype=np.float32)
print(f"{angles2.shape[0]} projections of images with dimension {projections2.shape[1:]} pixels")
print(f"{angles2.shape[0]} sets of {angles2.shape[1]} ground truth angles of corresponding projection images")
5000 projections of images with dimension (116, 116) pixels
5000 sets of 3 ground truth angles of corresponding projection images
plot_angles_histogram([angles2], plot_settings=dict(figsize=(10, 4)))
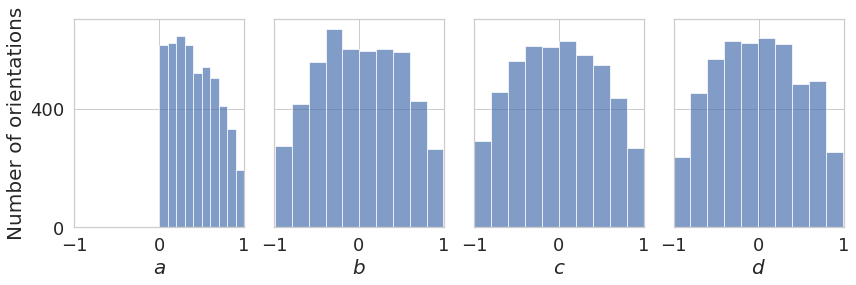
plot_quaternions_histogram([euler2quaternion(angles2)])
# pid = 0
# plot_projection(projections2[pid], f'Projection {pid}\nAngles {angles2[pid]}')
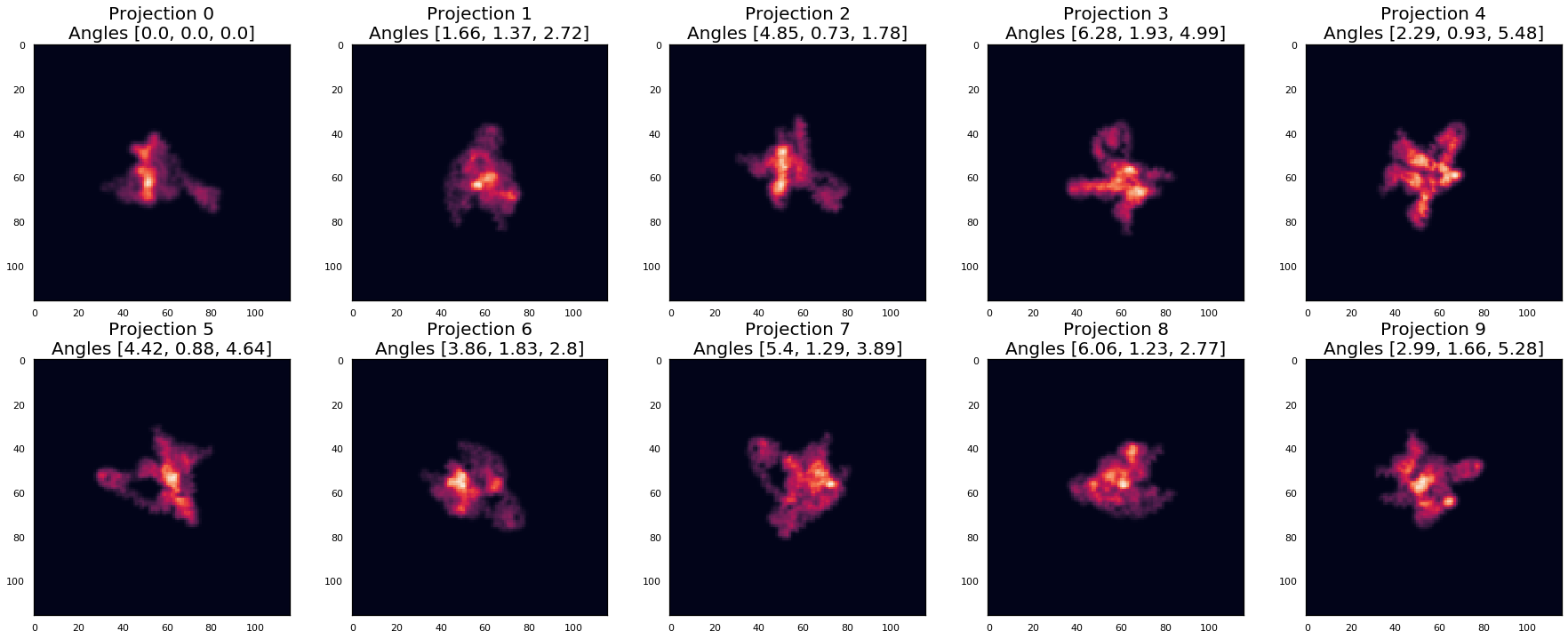
pids = range(10)
plot_projections(projections2[pids], [f'Projection {pid}\nAngles {list(map(lambda x: round(x,2) , angles2[pid]))}' for pid in pids], nrows=2, ncols=5)
plot_detector_pixels_with_protein(angles2, "../../data/5j0n.mrc", center=[34.88, 31.55, 39.82])
/home/jelena/miniconda3/envs/protein_reconstruction/lib/python3.6/site-packages/ipyvolume/serialize.py:81: RuntimeWarning: invalid value encountered in true_divide
gradient = gradient / np.sqrt(gradient[0]**2 + gradient[1]**2 + gradient[2]**2)
plot_images(angles2, projections2, indices=range(50), img_size_scale=0.15)
plot_rays(angles2, indices=range(50))